-
Notifications
You must be signed in to change notification settings - Fork 7
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Update from feeds: https://galaxyproject.org/news/2024-07-26-shiny-phyloseq-on-eu/ #92
base: main
Are you sure you want to change the base?
Conversation
…ical outcome of the Bioconductor Hackathon
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
This comment has been minimized.
|
|
||
| Shiny phyloseq interactive tool (IT) | ||
| ==================================== | ||
|
|
||
|
|
||
|  |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Those links unfortunately don't work, I guess we need to fix the RSS feed.
|
👋 Hello! I'm your friendly social media assistant. Below are the previews of this post: mastodon-eu-freiburg📝 New blog post Released! Shiny phyloseq interactive tool (IT) Shiny apps: https://shiny.posit.co/ are web apps using R functionality, that give easy responsive access to R packages. In theory all shiny apps could become ITs, but so far wrapping and deployment of shiny apps as ITs was technically challenging. To facilitate the process, the Freiburg Galaxy team conducted a two-day hackathon in February, collaborating with members of the Bioconductor community, including Charlotte Soneson, Hans-Rudolf Hotz, and Federico Marini. (3/12) During the event, best practices for developing ITs using Shiny apps were established, with a focus on creating a Docker image that can serve as a starting point for adding any Shiny app and the shiny-phyloseq app: https://github.com/joey711/shiny-phyloseq as a proof-of-concept target. (4/12) A fork of this Docker image tailored for the shiny app is available here: https://github.com/paulzierep/docker-phyloseq. This app allows to perform dynamic analysis of
The tool is available on usegalaxy.eu (https://usegalaxy.eu/?tool_id\=interactive_tool_phyloseq\&version\=latest) and was integrated into a dada2 based GTN tutorial: https://training.galaxyproject.org/training-material/topics/microbiome/tutorials/dada-16S/tutorial.html by Bérénice Batut. (7/12) Data upload from Galaxy The original shiny-phyloseq app worked in an IT, but did not allow to upload amplicon data via CLI arguments making data input from Galaxy incovenient (requiring users to download data from their Galaxy History, then upload it to the IT via the web frontend). (8/12) Therefore, the tool was forked and modified to allow for additional data input. This functional adaptation of shiny-apps to allow for non-web based data upload is the only requirement to make any shiny app compatible with Galaxy based data upload. (9/12) The changes made to the original app can be found in this git diff: joey711/shiny-phyloseq@master...paulzierep:shiny-phyloseq:master. (10/12) Outlook To improve the usability and Galaxy-interaction of the phyloseq-shiny app we are working on the possibility to export data (figures, modified phyloseq objects) to the Galaxy history. (11/12) This could be done either by collecting the data as outputs of the IT or by using utility functions such as put / get, that are used in other ITs like RStudio: https://usegalaxy.eu/?tool_id=interactive_tool_rstudio&version=latest.
bluesky-galaxyproject📝 New blog post Released! Shiny phyloseq interactive tool (IT) Shiny apps: https://shiny.posit.co/ are web apps using R functionality, that give easy responsive access to R packages. In theory all shiny apps could become ITs, but so far wrapping and deployment of shiny apps as ITs was technically challenging. To facilitate the process, the Freiburg Galaxy team conducted a two-day hackathon in February, collaborating with members of the Bioconductor community, including (3/14) Charlotte Soneson, Hans-Rudolf Hotz, and Federico Marini. (4/14) During the event, best practices for developing ITs using Shiny apps were established, with a focus on creating a Docker image that can serve as a starting point for adding any Shiny app and the shiny-phyloseq app: https://github.com/joey711/shiny-phyloseq as a proof-of-concept target. (5/14) A fork of this Docker image tailored for the shiny app is available here: https://github.com/paulzierep/docker-phyloseq. This app allows to perform dynamic analysis of
The tool is available on usegalaxy.eu (https://usegalaxy.eu/?tool_id\=interactive_tool_phyloseq\&version\=latest) and was integrated into a dada2 based GTN tutorial: https://training.galaxyproject.org/training-material/topics/microbiome/tutorials/dada-16S/tutorial.html by Bérénice Batut. (8/14) Data upload from Galaxy The original shiny-phyloseq app worked in an IT, but did not allow to upload amplicon data via CLI arguments making data input from Galaxy incovenient (requiring users to download data from their Galaxy History, then upload it to the IT via the web frontend). (9/14) Therefore, the tool was forked and modified to allow for additional data input. This functional adaptation of shiny-apps to allow for non-web based data upload is the only requirement to make any shiny app compatible with Galaxy based data upload. (10/14) The changes made to the original app can be found in this git diff: joey711/shiny-phyloseq@master...paulzierep:shiny-phyloseq:master. (11/14) Outlook To improve the usability and Galaxy-interaction of the phyloseq-shiny app we are working on the possibility to export data (figures, modified phyloseq objects) to the Galaxy history. (12/14) This could be done either by collecting the data as outputs of the IT or by using utility functions such as put / get, that are used in other ITs like RStudio: https://usegalaxy.eu/?tool_id=interactive_tool_rstudio&version=latest.
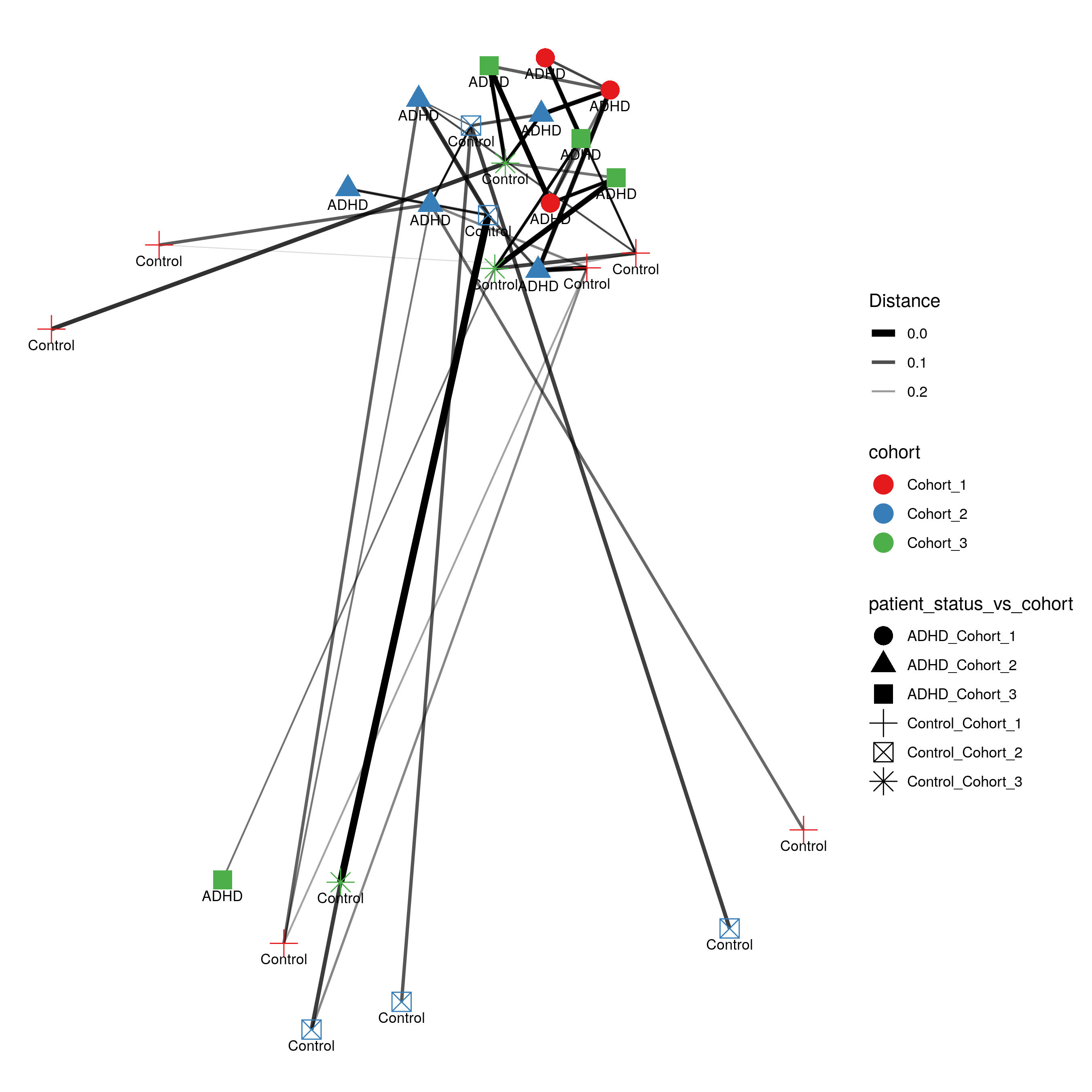
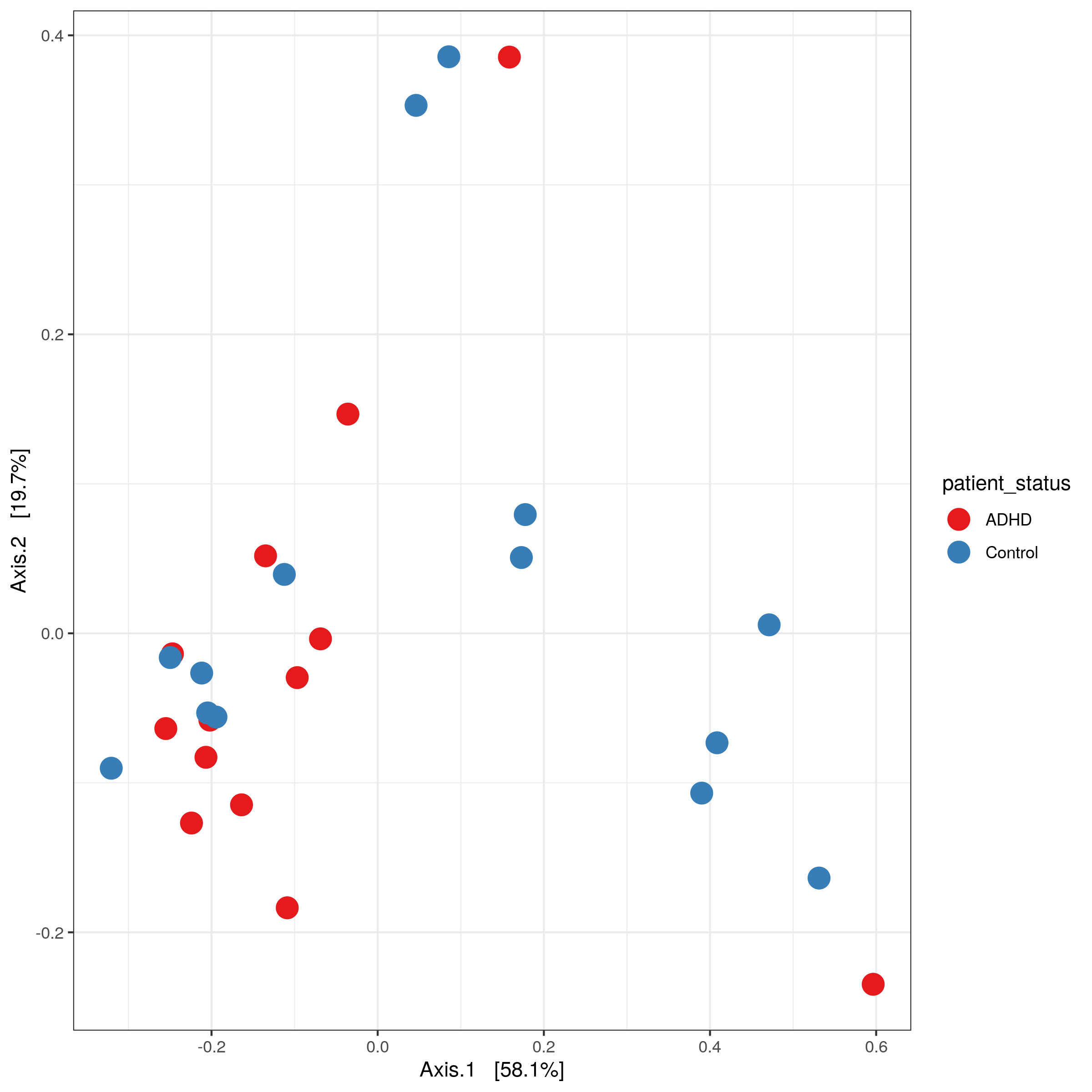
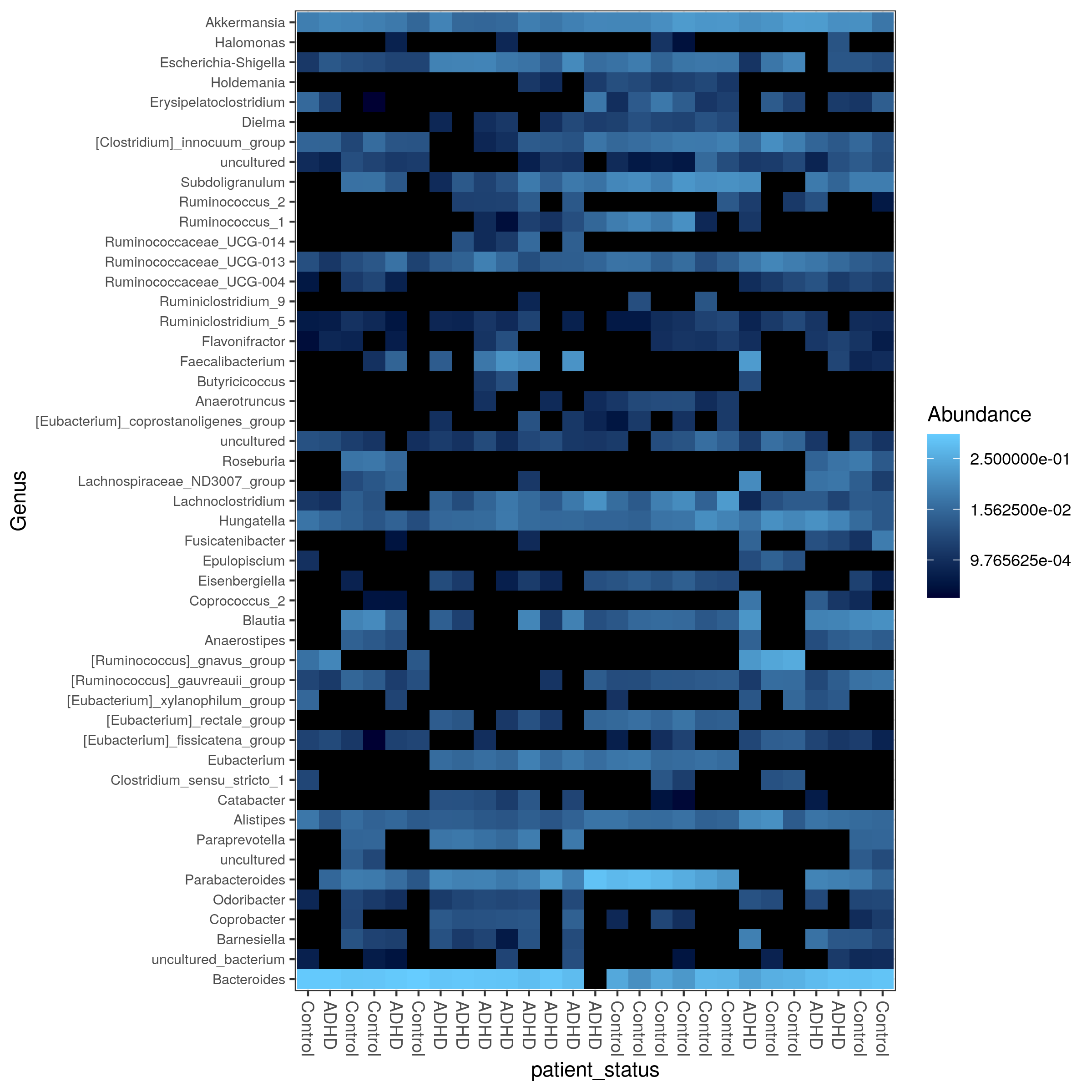
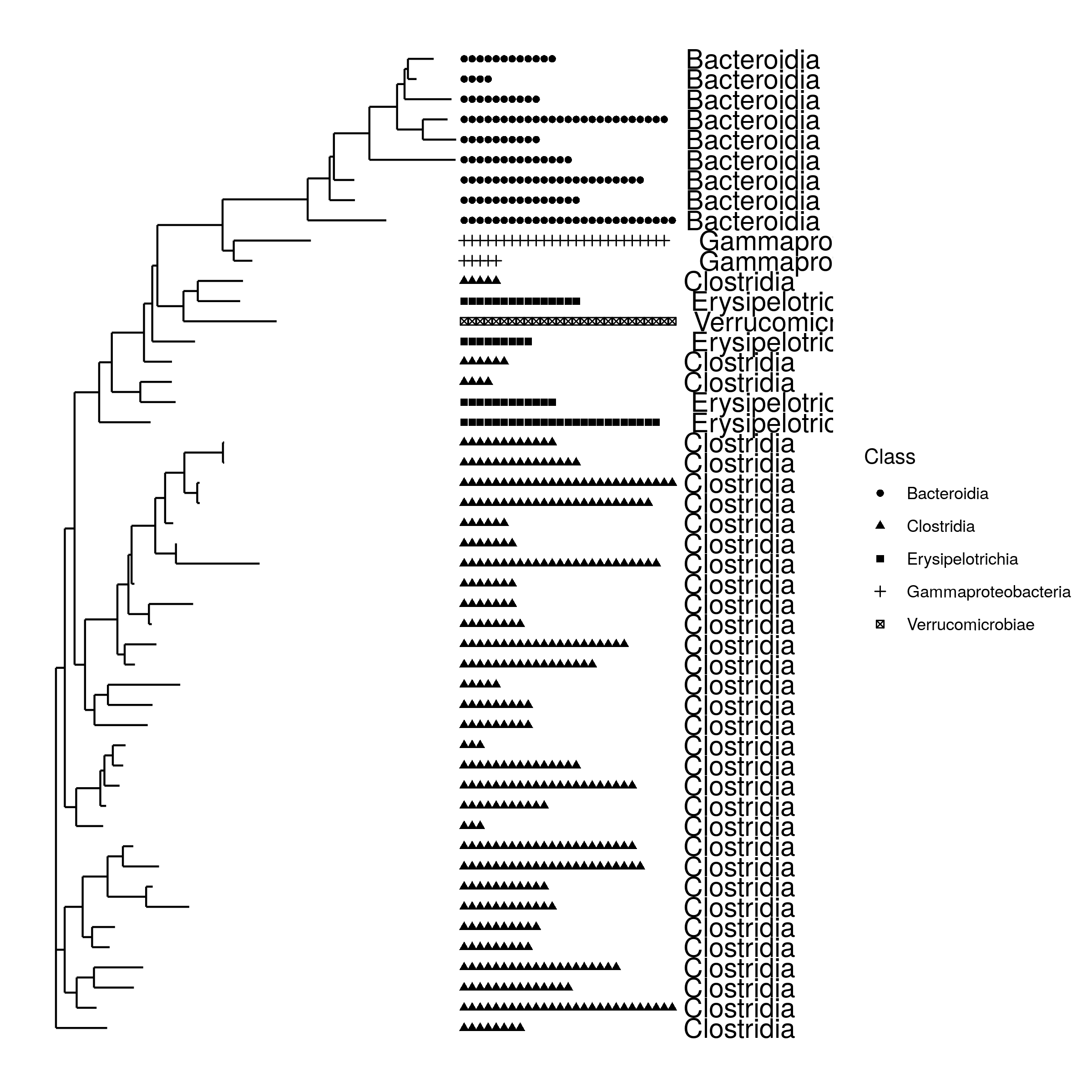
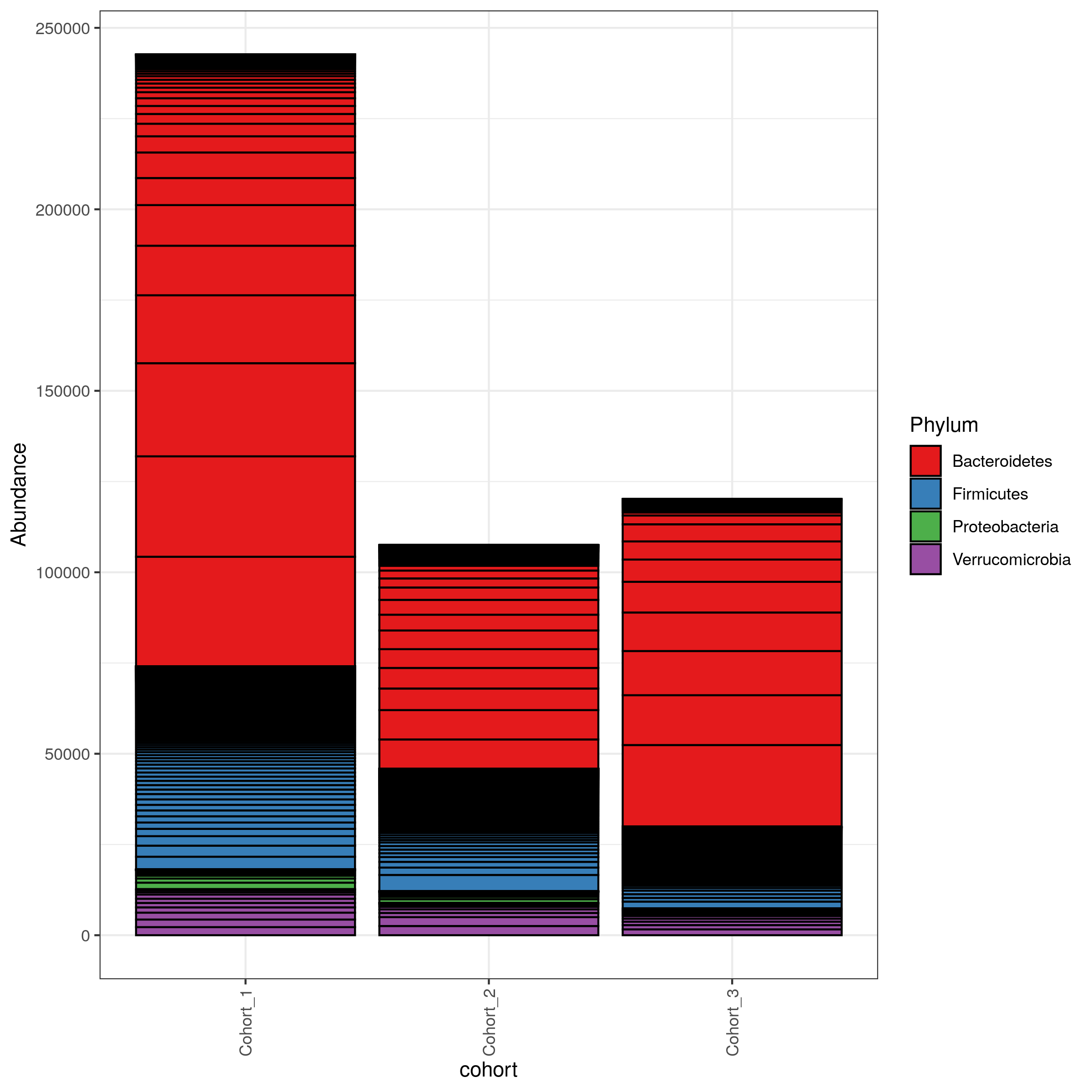
matrix-eu-announcebgruening: 📝 New blog post Released! https://galaxyproject.org/news/2024-07-26-shiny-phyloseq-on-eu/ Shiny phyloseq interactive tool (IT)Shiny apps are web apps using R functionality, that give easy responsive access to R packages. Interactive tools are a great way to work with data interactively and responsive using Galaxy. In theory all shiny apps could become ITs, but so far wrapping and deployment of shiny apps as ITs was technically challenging. To facilitate the process, the Freiburg Galaxy team conducted a two-day hackathon in February, collaborating with members of the Bioconductor community, including Charlotte Soneson, Hans-Rudolf Hotz, and Federico Marini. During the event, best practices for developing ITs using Shiny apps were established, with a focus on creating a Docker image that can serve as a starting point for adding any Shiny app and the shiny-phyloseq app as a proof-of-concept target. A fork of this Docker image tailored for the shiny app is available here: https://github.com/paulzierep/docker-phyloseq. The image can be run locally to test the app and then must be deployed to quay.io or any other Docker registry. This app allows to perform dynamic analysis of metabarcoding/amplicon data such as:
The tool is available on usegalaxy.eu (https://usegalaxy.eu/?tool_id\=interactive_tool_phyloseq\&version\=latest) and was integrated into a dada2 based GTN tutorial by Bérénice Batut. Data upload from GalaxyThe original shiny-phyloseq app worked in an IT, but did not allow to upload amplicon data via CLI arguments making data input from Galaxy incovenient (requiring users to download data from their Galaxy History, then upload it to the IT via the web frontend). Therefore, the tool was forked and modified to allow for additional data input. This functional adaptation of shiny-apps to allow for non-web based data upload is the only requirement to make any shiny app compatible with Galaxy based data upload. The changes made to the original app can be found in this git diff. OutlookTo improve the usability and Galaxy-interaction of the phyloseq-shiny app we are working on the possibility to export data (figures, modified phyloseq objects) to the Galaxy history. This could be done either by collecting the data as outputs of the IT or by using utility functions such as put / get, that are used in other ITs like RStudio. linkedin-galaxyproject📝 New blog post Released! Shiny phyloseq interactive tool (IT) Shiny apps: https://shiny.posit.co/ are web apps using R functionality, that give easy responsive access to R packages. In theory all shiny apps could become ITs, but so far wrapping and deployment of shiny apps as ITs was technically challenging. To facilitate the process, the Freiburg Galaxy team conducted a two-day hackathon in February, collaborating with members of the Bioconductor community, including Charlotte Soneson, Hans-Rudolf Hotz, and Federico Marini. During the event, best practices for developing ITs using Shiny apps were established, with a focus on creating a Docker image that can serve as a starting point for adding any Shiny app and the shiny-phyloseq app: https://github.com/joey711/shiny-phyloseq as a proof-of-concept target. A fork of this Docker image tailored for the shiny app is available here: https://github.com/paulzierep/docker-phyloseq. This app allows to perform dynamic analysis of
The tool is available on usegalaxy.eu (https://usegalaxy.eu/?tool_id\=interactive_tool_phyloseq\&version\=latest) and was integrated into a dada2 based GTN tutorial: https://training.galaxyproject.org/training-material/topics/microbiome/tutorials/dada-16S/tutorial.html by Bérénice Batut. Data upload from Galaxy The original shiny-phyloseq app worked in an IT, but did not allow to upload amplicon data via CLI arguments making data input from Galaxy incovenient (requiring users to download data from their Galaxy History, then upload it to the IT via the web frontend). Therefore, the tool was forked and modified to allow for additional data input. This functional adaptation of shiny-apps to allow for non-web based data upload is the only requirement to make any shiny app compatible with Galaxy based data upload. The changes made to the original app can be found in this git diff: joey711/shiny-phyloseq@master...paulzierep:shiny-phyloseq:master. Outlook To improve the usability and Galaxy-interaction of the phyloseq-shiny app we are working on the possibility to export data (figures, modified phyloseq objects) to the Galaxy history. This could be done either by collecting the data as outputs of the IT or by using utility functions such as put / get, that are used in other ITs like RStudio: https://usegalaxy.eu/?tool_id=interactive_tool_rstudio&version=latest.
|
|
Observed problems with generating the posts from the feed:
|
|
In addition, all the image links generated for the preview are broken. |
|
and in general, supporting bluesky (with its super-low per post char limit) for the same content as the other platforms is a pain forcing us to split the content into much smaller chunks than what would be needed for the other platforms. Of course, maintaining extra posts to bluesky is also painful, but I don't have a better solution. Any ideas? |
|
For the bot it does not matter if we create a bluesky post separately. We could do that. |
Yes, but for making it look pretty it would still require manual insertion of paragraphs (so extra work). |
|
@wm75 The images are now fixed thanks to Dannon and the rest of the bugs have been addressed in usegalaxy-eu/galaxy-social-assistant@ad496e5. |

This PR is created automatically by a feeds bot.
Update since 2024-07-26
Processed:
Shiny phyloseq interactive tool available on usegalaxy.eu - practical outcome of the Bioconductor Hackathon