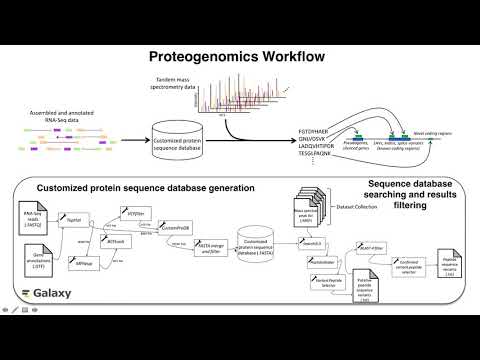
This page provides links to instructional materials describing how to access and use the proteogenomic workflows described in the manuscript text.
Please access the links indicated below for the following materials:
**Training on usegalaxyp.org**
- How to access and use the usegalaxyp.org training site: http://z.umn.edu/accessusegalaxyporg
- Mouse Protegenomics tutorial: http://z.umn.edu/pginnov18
**Interactive Galaxy Tours**
-
Accessing an interactive Galaxy Tour describing the RNA-seq-to-protein-database workflow: http://z.umn.edu/rnaseqtoproteindbtour
-
Accessing an interactive Galaxy Tour describing Dataset Collections: http://z.umn.edu/datasetcollectiontour
-
Accessing an interactive Galaxy Tour describing the protein sequence database search workflow: http://z.umn.edu/sequencedbsearchtour
**Workflow training and operation**
-
Instructions on running the RNA-seq-to-protein-database workflow: http://z.umn.edu/rnatoproteindbworkflow
-
Instructions on running the protein sequence database search workflow and interpreting results: http://z.umn.edu/sequencedbsearchworkflow
**JetStream instance**
- Instructions for accessing workflows on the JetStream Galaxy instance: http://z.umn.edu/accessjetstream