Created at the University of Toronto and published in Oxford Bioinformatics (2016, 2023).
Authored by: Max Franz, Christian Lopes, Dylan Fong, Mike Kucera, ..., Gary Bader
Graph theory (network) library for visualisation and analysis : https://js.cytoscape.org
Cytoscape.js is a fully featured graph theory library. Do you need to model and/or visualise relational data, like biological data or social networks? If so, Cytoscape.js is just what you need.
Cytoscape.js contains a graph theory model and an optional renderer to display interactive graphs. This library was designed to make it as easy as possible for programmers and scientists to use graph theory in their apps, whether it's for server-side analysis in a Node.js app or for a rich user interface.
You can get started with Cytoscape.js with one line:
var cy = cytoscape({ elements: myElements, container: myDiv });Learn more about the features of Cytoscape.js by reading its documentation.
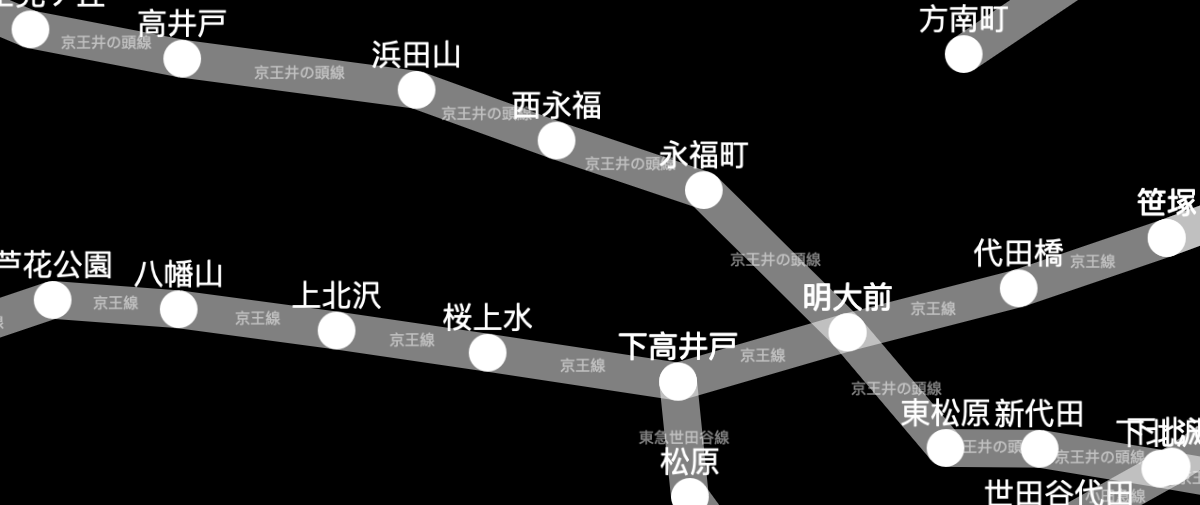
The Tokyo railway stations network can be visualised with Cytoscape:
A live demo and source code are available for the Tokyo railway stations graph. More demos are available in the documentation.
You can find the documentation and downloads on the project website.
Future versions of Cytoscape.js are planned in the milestones of the Github issue tracker. You can use the milestones to see what's currently planned for future releases.
Would you like to become a Cytoscape.js contributor? You can contribute in technical roles (e.g. features, testing) or non-technical roles (e.g. documentation, outreach), depending on your interests. Get in touch with us by posting a GitHub discussion.
For the mechanics of contributing a pull request, refer to CONTRIBUTING.md.
Feature releases are made monthly, while patch releases are made weekly. This allows for rapid releases of first- and third-party contributions.
To cite Cytoscape.js in a paper, please cite the Oxford Bioinformatics issue:
Cytoscape.js: a graph theory library for visualisation and analysis
Franz M, Lopes CT, Huck G, Dong Y, Sumer O, Bader GD
Bioinformatics (2016) 32 (2): 309-311 first published online September 28, 2015 doi:10.1093/bioinformatics/btv557 (PDF)
Install node and npm. Run npm install before using npm run.
Run npm run <target> in the console. The main targets are:
Building:
build: do all builds of the library (umd, min, umd, esm)build:min: do the unminified build with bundled dependencies (for simple html pages, good for novices)build:umd: do the umd (cjs/amd/globals) buildbuild:esm: do the esm (ES 2015 modules) buildclean: clean thebuilddirectorydocs: build the docs intodocumentationrelease: build all release artifactswatch: automatically build lib for debugging (with sourcemap, no babel, very quick)- good for general testing on
debug/index.html - served on
http://localhost:8080or the first available port thereafter, with livereload ondebug/index.html
- good for general testing on
watch:babel: automatically build lib for debugging (with sourcemap, with babel, a bit slower)- good for testing performance or for testing out of date browsers
- served on
http://localhost:8080or the first available port thereafter, with livereload ondebug/index.html
watch:umd: automatically build prod umd bundle (no sourcemap, with babel)- good for testing cytoscape in another project (with a
"cytoscape": "file:./path/to/cytoscape"reference in your project'spackage.json) - no http server
- good for testing cytoscape in another project (with a
dist: update the distribution js for npm etc.
Testing:
The default test scripts run directly against the source code. Tests can alternatively be run on a built bundle. The library can be built on node>=6, but the library's bundle can be tested on node>=0.10.
test: run all testing & lintingtest:js: run the mocha tests on the public API of the lib (directly on source files)npm run test:js -- -g "my test name"runs tests on only the matching test cases
test:build: run the mocha tests on the public API of the lib (on a built bundle)npm run buildshould be run beforehand on a recent version of nodenpm run test:build -- -g "my test name"runs build tests on only the matching test cases
test:modules: run unit tests on private, internal APInpm run test:modules -- -g "my test name"runs modules tests on only the matching test cases
lint: lint the js sources via eslintbenchmark: run all benchmarksbenchmark:single: run benchmarks only for the suite specified inbenchmark/single
- Ensure that a milestone exists for the release you want to make, with all the issues for that release assigned in the milestone.
- Bug fixes should be applied to both the
masterandunstablebranches. PRs can go on either branch, with the patch applied to the other branch after merging. - When a patch release is made concurrently with a feature release, the patch release should be made first. Wait 5 minutes after the patch release completes before starting the feature release -- otherwise Zenodo doesn't pick up releases properly.
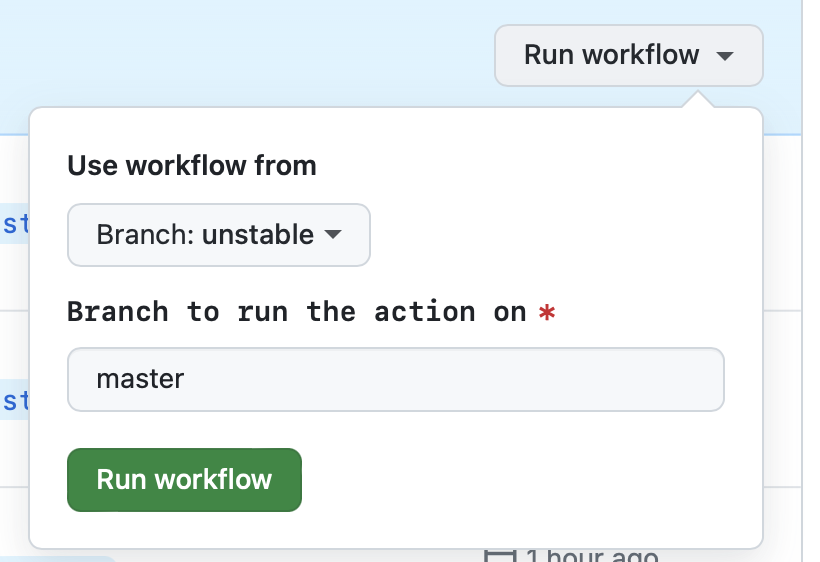
- Go to Actions > Patch release
- Go to the 'Run workflow' dropdown
- [Optional] The 'master' branch should be preselected for you
- Press the green 'Run workflow' button
- Close the milestone for the release
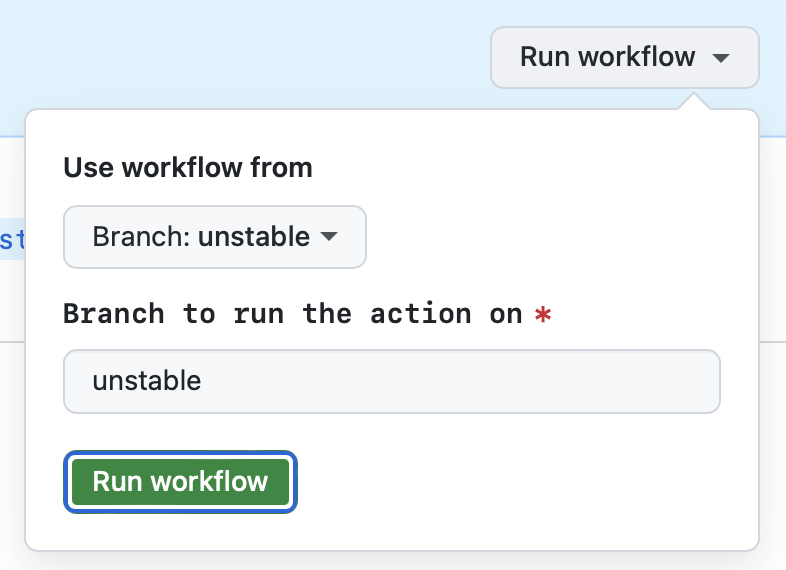
- Go to Actions > Feature release
- Go to the 'Run workflow' dropdown
- [Optional] The 'unstable' branch should be preselected for you
- Press the green 'Run workflow' button
- Close the milestone for the release
- Make the release announcement on the blog
- 'Use workflow from' in the GitHub UI selects the branch from which the workflow YML file is selected. Since the workflow files should usually be the same on the master and unstable branches, it shouldn't matter what's selected.
- 'Branch to run the action on' in the GitHub UI is preselected for you. You don't need to change it.
Mocha tests are found in the test directory. The tests can be run in the browser or they can be run via Node.js (npm run test:js).